Please visit www.templateflow.org for a more comprehensive description of this project. News and some discussions take place at the Nipy discourse platform.
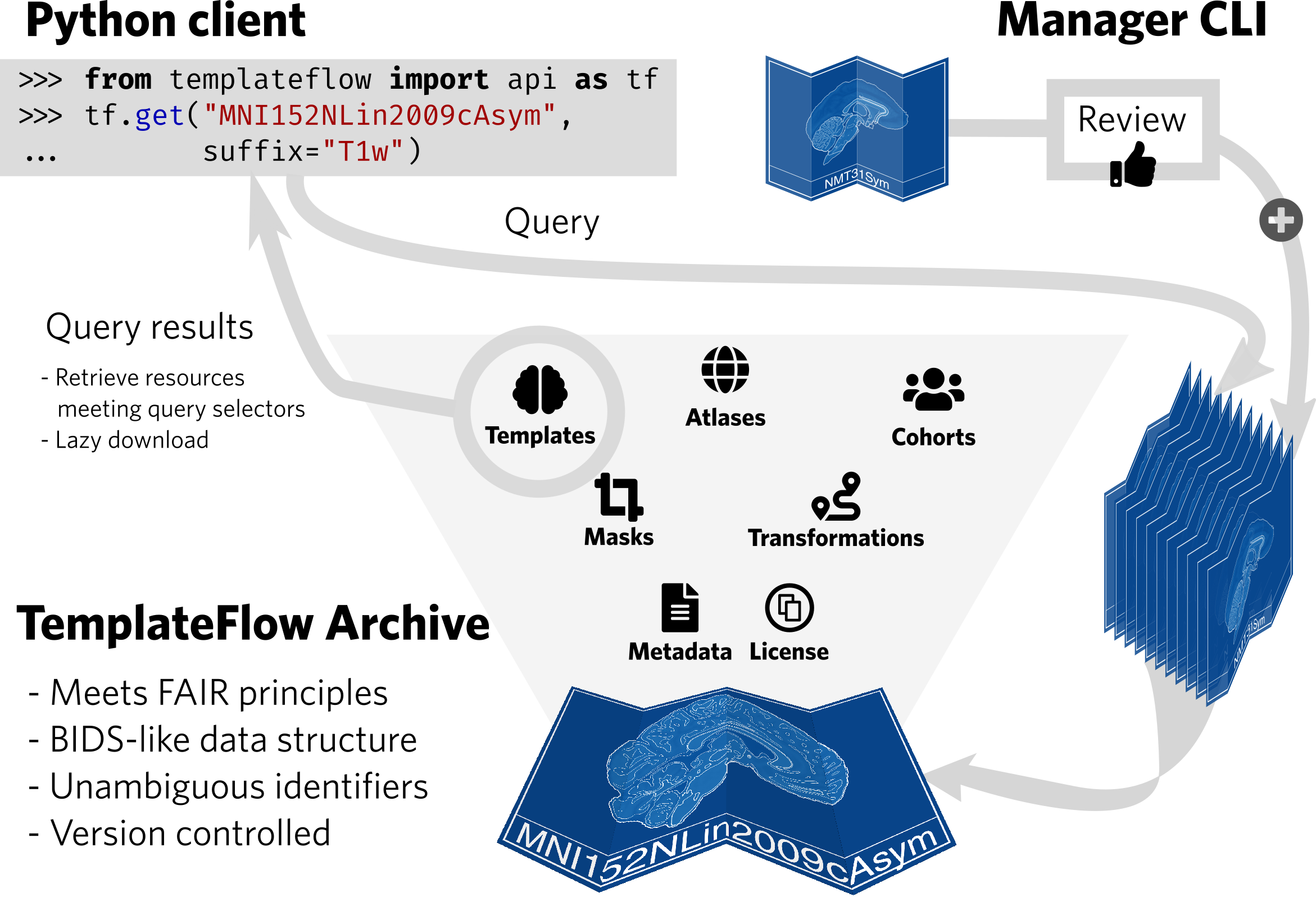
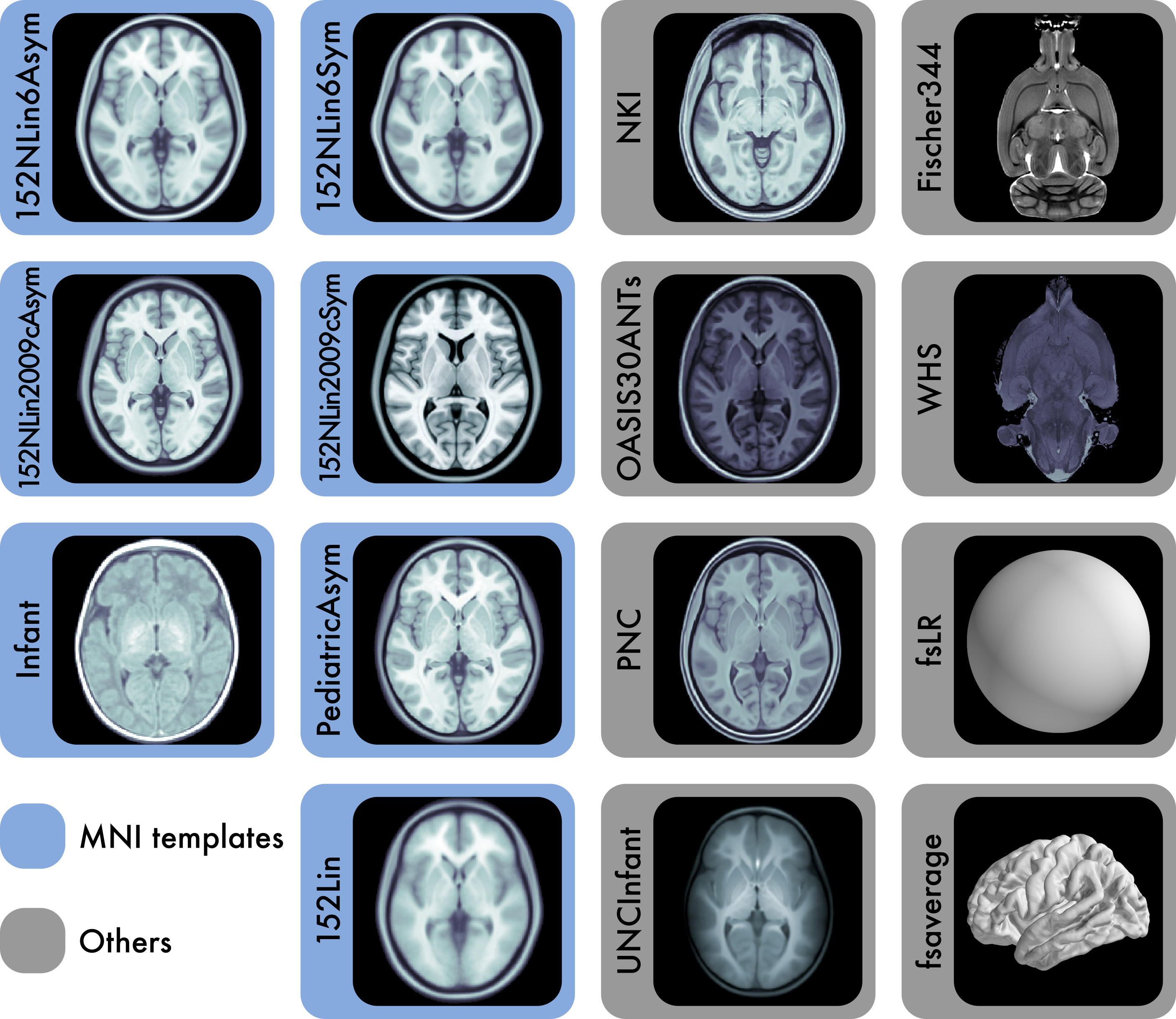
Reference anatomies of the brain and corresponding atlases play a central role in experimental neuroimaging workflows and are the foundation for reporting standardized results. The choice of such references —i.e., templates— and atlases is one relevant source of methodological variability across studies, which has recently been brought to attention as an important challenge to reproducibility in neuroscience. TemplateFlow is a publicly available framework for human and nonhuman brain models. The framework combines an open database with software for access, management, and vetting, allowing scientists to distribute their resources under FAIR —findable, accessible, interoperable, reusable— principles. TemplateFlow supports a multifaceted insight into brains across species and enables multiverse analyses testing whether results generalize across standard references, scales, and in the long term, species, thereby contributing to increasing the reliability of neuroimaging results.
The rationale behind TemplateFlow and how we envision it as a fundamental instrument for neuroimaging studies is presented in our preprint:
Ciric, R., Thompson, W.H., Lorenz, R. et al. TemplateFlow: FAIR-sharing of multi-scale, multi-species brain models. Nat Methods 19, 1568–1571 (2022). doi:10.1038/s41592-022-01681-2
An extended version of this paper is found in our pre-print.
This work is steered and maintained by the NiPreps Community. The development of this framework is supported the NIMH (RF1MH121867, RAP, OE).
Thanks to the DataLad developers, as we rely on their excellent tool for managing the TemplateFlow Archive.