| Health | Main Release (w/ Mantid Framework) | Dev Release (w/o Mantid Framework) |
|---|---|---|
 |
||
This project is trying to implement total scattering data reduction for neutron time-of-flight diffractometers using the algorithms currently available in the Mantid framework
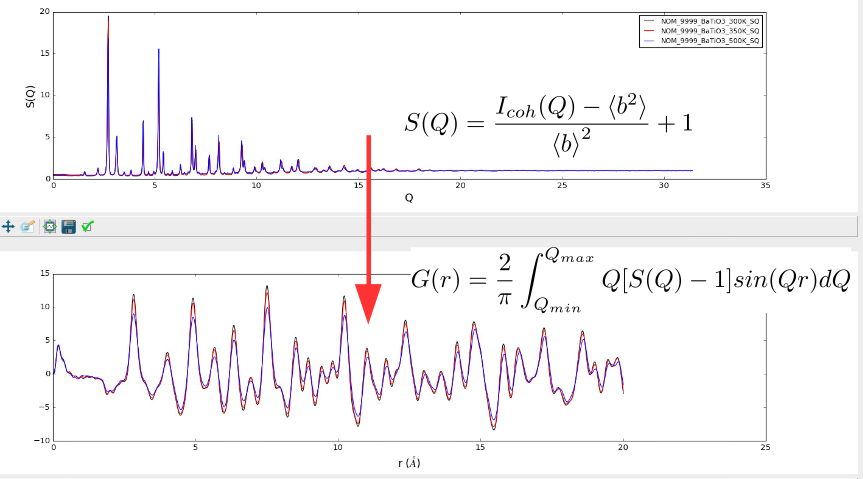
This entails taking raw neutron counts from detectors in the diffraction experiment and turning them into the reciprocal-space structure factor patterns, F(Q) or S(Q), and applying a Fourier Transform to real-space to give the pair distribution fuction, PDF.
This is the future backend for the ADDIE project and hopes to support multiple diffractometers performing total scattering measurements.
Running mantidtotalscattering will generate the total scattering data in
reciprocal space saved as NeXus file, if one is using the multiple bank mode.
Another alternative mode is the single bank mode which will merge spectra from
all detectors into a single pattern. The single bank mode will be taken in the
auto reduction implementation since otherwise manual efforts are needed to merge
the different banks data. When using the single bank mode, no Bragg data will be
saved. With the multiple bank mode, both the total scattering data and the Bragg
diffraction data will be saved. For the Bragg data output, there are two
available formats -- the GSS format and the XYE format. The GSS data file could
be loaded into the ADDIE interface (see the link above) for visualization. Here,
it is noteworthy that if one is loading in the GSS data in Mantid, one has to
rebin the loaded in workspace first (since the output GSS data is ragged, i.e.,
different banks are not sharing the common x-axis, for the purpose of removing
the non-sense data in each bank), followed by running the Mantid algorithm
ConvertToPointData.
The following commands can get you setup (on Linux machine) to get conda installed (as miniconda):
CONDA_PYTHON=3
MINICONDA_URL="https://repo.continuum.io/miniconda";
MINICONDA_FILE="Miniconda${CONDA_PYTHON}-latest-Linux-x86_64.sh";
wget "${MINICONDA_URL}/${MINICONDA_FILE}";
bash ${MINICONDA_FILE} -b -p $HOME/miniconda;
export PATH="$HOME/miniconda/bin:$PATH";
NOTE: You can change the python version number via the CONDA_PYTHON variable
You will have to excute the last command on every new bash session (export PATH...).
Adding this last line to your ~/.bashrc will automatically add it on every bash session startup.
Add channels with dependencies, create a conda environment with python_version set to either 2.7.14 or 3.6, and activate the environment
conda config --add channels conda-forge --add channels mantid --add channels mantid/label/nightly
conda create -n mantidts_env python=${python_version}
source activate mantidts_envNOTE: Mamba is still in "beta".
python_version=3.6
conda config --add channels conda-forge --add channels mantid --add channels mantid/label/nightly
conda install mamba -c conda-forge
mamba update mamba -c conda-forge
mamba create -n mantidts_env python=${python_version}
source activate mantidts_env
Simply replace conda -> mamba in "Install" instruction commands
conda install -c neutrons mantid-total-scatteringGo here for how to delete an environment or use:
conda remove --name mantidts_env --allIf you have an error (see below for example) related to the libGL library, you may not have it installed for the Mantid Framework to work. See instructions here for installing the necessary libraries for different OS
Example error:
ImportError: First import of "._api" failed with "libGL.so.1: cannot open shared object file...
If you have an error that another version of Mantid is installed on the machine and being imported via PYTHONPATH, you can use the following as a workaround for CLI tool:
PYTHONPATH="" mantidtotalscatteringTo launch the total scattering script, complete the input JSON file (found in examples directory), and run:
mantidtotalscattering examples/sns/nomad_simple.jsonIf you need to specify the path to Mantid build, use:
MANTIDPATH=/path/to/mantid/build/bin PATH=$MANTIDPATH:$PATH PYTHONPATH=$MANTIDPATH:$PATH mantidtotalscattering <json input>This is mainly for development if you want to use a local development build of Mantid Framework instead of one included.
pip install mantid-total-scatteringAdd channels with dependencies, create a conda environment with python_version set to either 2.7 or 3.6, and activate the environment
conda config --add channels conda-forge
conda create -n mantidts_env python=${python_version}
conda activate mantidts_envconda install -c neutrons mantid-total-scattering-python-wrapperClone the repository to a local directory
git clone https://github.com/neutrons/mantid_total_scattering.git
cd mantid_total_scatteringLocated in the main directory of the repo, we have several ways to do the local
testing for mantidtotalscattering using a selected version of mantid build.
The new way of Mantid building has been changed to use conda, and detailed
information can be found here, https://developer.mantidproject.org/GettingStarted/GettingStartedCondaLinux.html#gettingstartedcondalinux.
Suppose we are following exactly the instruction in the link above to build
Mantid, we will have a conda environment mantid-developer, in which case we
can execute the command below to use the local Mantid build to work with the
local version of mantidtotalscattering,
conda activate mantid-developer
python MANTID_REPO_DIR/build/bin/AddPythonPath.py
python total_scattering/cli.py INPUT_JSON_FILEwhere MANTID_REPO_DIR refers to the full path of the mantid repo directory.
The second way we can try is to launch mantidtotalscattering from within
mantidworkbench. To do this, again, assuming we are located in the main
mantidtotalscattering directory, we can execute,
MANTID_REPO_DIR/build/bin/launch_mantidworkbench.shto start up the local version of mantidworkbench. Then we can load the
following script into mantidworkbench and execute it,
import json
from total_scattering.reduction import TotalScatteringReduction
from total_scattering.reduction import validateConfig
with open("FULL_PATH_TO_INPUT_JSON_FILE", "r") as handle:
config = json.load(handle)
# validate the config
validateConfig(config)
# Run total scattering reduction
TotalScatteringReduction(config)where we need to replace FULL_PATH_TO_INPUT_JSON_FILE with the full path to
our input json file.
N. B. To build a local version of mantid framework, we can check out
this link, https://developer.mantidproject.org/GettingStarted/GettingStartedCondaLinux.html#gettingstartedcondalinux.
The third way is to set up a local virtual environment for
mantidtotalscattering and add in the python path of local Mantid build. To do
this, follow the steps below,
-
virtualenv -p MANTID-DEVELOPER_ENV_LOCATION/bin/python --system-site-packages .venv
-
source .venv/bin/activate
-
python MANTID_REPO_DIR/build/bin/AddPythonPath.py
-
pip install -r requirements.txt -r requirements-dev.txt
-
python setup.py develop
where MANTID_REPO_DIR refers to the full path of the mantid repo directory.
N.B. With the third way of setup, one could then execute
python SCRIPT_NAME.pyto runmantidtotalscatteringreduction. The script can be with the contents as shared above. Meanwhile, one can also executemantidpython SCRIPT_NAME.pyif on ORNL analysis cluster. One interesting observation is if using the former way (i.e., runpython SCRIPT_NAME.py), theSavePlot1Dalgorithm cannot take theOutputTypeas beingplotly-all, whereas using the latter command will be able to do that.
To build and run the tests via pytest, use:
python setup.py testN. B. This is assuming that the mantid-developer conda environment mentioned
above is active.
To build and run tests via Docker, use:
docker build -t unit-test-env -f .ci/Dockerfile.nightly_ubuntu16.04_python3 . && docker run -t unit-test-env /bin/bash -c "mantidpython -m pytest"Mantid Total Scattering uses versioneer. These are the instructions to create a new version, working on a local clone,
git branch --track main origin/main # create a local main branch set to follow remote main
git checkout main
git fetch -p -t # fetch all changes from the remote repo
git rebase -v origin/main # sync with remote main branch
git merge --ff-only origin/qa # merge in all of the changes in branch next
git tag v1.0.7 # create the tag in the format that versioneer has been configured
git push origin v1.0.7 # push the tag to remote to kick off the deploy stepThe steps above will tag a new main release (the production release) and will
kick off the pipeline action which will further trigger the connected GitLab
action to deploy the built conda package to SNS analysis cluster (to the central
mantidtotalscattering conda environment located at /opt/anaconda/envs). If
we want to tag a release candidate, follow the steps below,
git branch --track qa origin/qa # create a local main branch set to follow remote main
git checkout qa
git fetch -p -t # fetch all changes from the remote repo
git rebase -v origin/qa # sync with remote main branch
git merge --ff-only origin/next # merge in all of the changes in branch next
git tag v1.0.8rc1 # create the tag in the format that versioneer has been configured
git push origin v1.0.8rc1 # push the tag to remote to kick off the deploy stepThis will kick off the pipeline action which will further trigger the connected
GitLab action to deploy the built conda package to SNS analysis cluster (to the
central mantidtotalscattering-qa conda environment located at /opt/anaconda/envs).