Atomic Force Microscopy (AFM) with a CO functionalized tip plays a crucial role in characterizing atomic-scale nanostructures. However, identifying structures from AFM images is a challenging task that relies heavily on human expertise. Simulations, particularly with Particle Probe AFM (PPAFM) [1], offer a cost-effective solution for generating large volumes of AFM images. By training state-of-the-art machine learning models with extensive PPAFM-generated datasets, the underlying molecular geometry can be predicted accurately.
In our previous works [2,3,4], we developed machine learning workflows based on a two-stage process. To enhance this workflow, we aim to use an end-to-end model, which simplifies the training process and improves overall efficiency. Such a model has great potential for application to experimental AFM images, enabling faster and more reliable structure discovery.
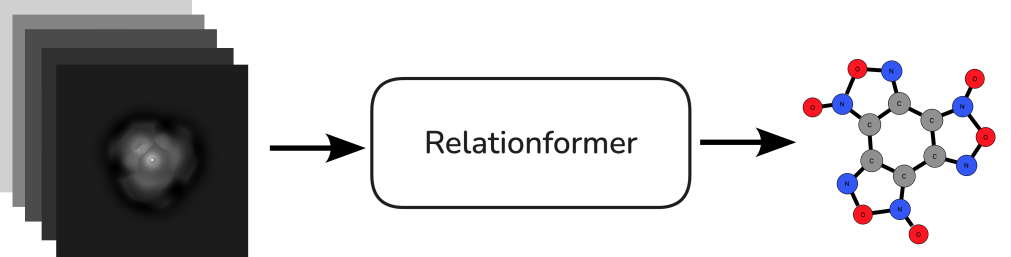
During the hackathon, we successfully started training an image-to-graph translation tool, Relationformer [5], with non-contact AFM (nc-AFM) images to predict sample structures. Specifically, we used a transformer-based model capable of simultaneously predicting objects (atoms) and their relationships (bonds), ensuring accurate geometric characterization. To train this model, we utilized a high-throughput nc-AFM simulator, PPAFM, which provides high-resolution AFM images alongside molecular graph labels.
- Simulation data: We have use PPM model to get the simulated images.
- Image-to-Graph model: We have use the Relationformer as our end-to-end model.
- Accurate prediction of molecular graphs, including atom types and bond information, from AFM images.
- Enhanced workflow efficiency by transitioning to an end-to-end machine learning model.
-
N. Oinonen, A. V. Yakutovich, A. Gallardo, M. Ondracek, P. Hapala, O. Krejci, Advancing Scanning Probe Microscopy Simulations: A Decade of Development in Probe-Particle Models, Comput. Phys. Commun. 305, 109341 (2024).
-
F. Priante, N. Oinonen, Y. Tian, D. Guan, C. Xu, S. Cai, P. Liljeroth, Y. Jiang, A. S. Foster, ACS Nano 18(7), 5546-5555 (2024).
-
L. Kurki, N. Oinonen, A. S. Foster, ACS Nano 18(17), 11130-11138 (2024).
-
N. Oinonen, L. Kurki, A. Ilin et al., Molecule Graph Reconstruction from Atomic Force Microscope Images with Machine Learning, MRS Bull. 47, 895–905 (2022).
-
S. Shit, R. Koner, B. Wittmann et al., Relationformer: A Unified Framework for Image-to-Graph Generation, arXiv:2203.10202 (2022).
We thank the authors of the project of Relationformer.