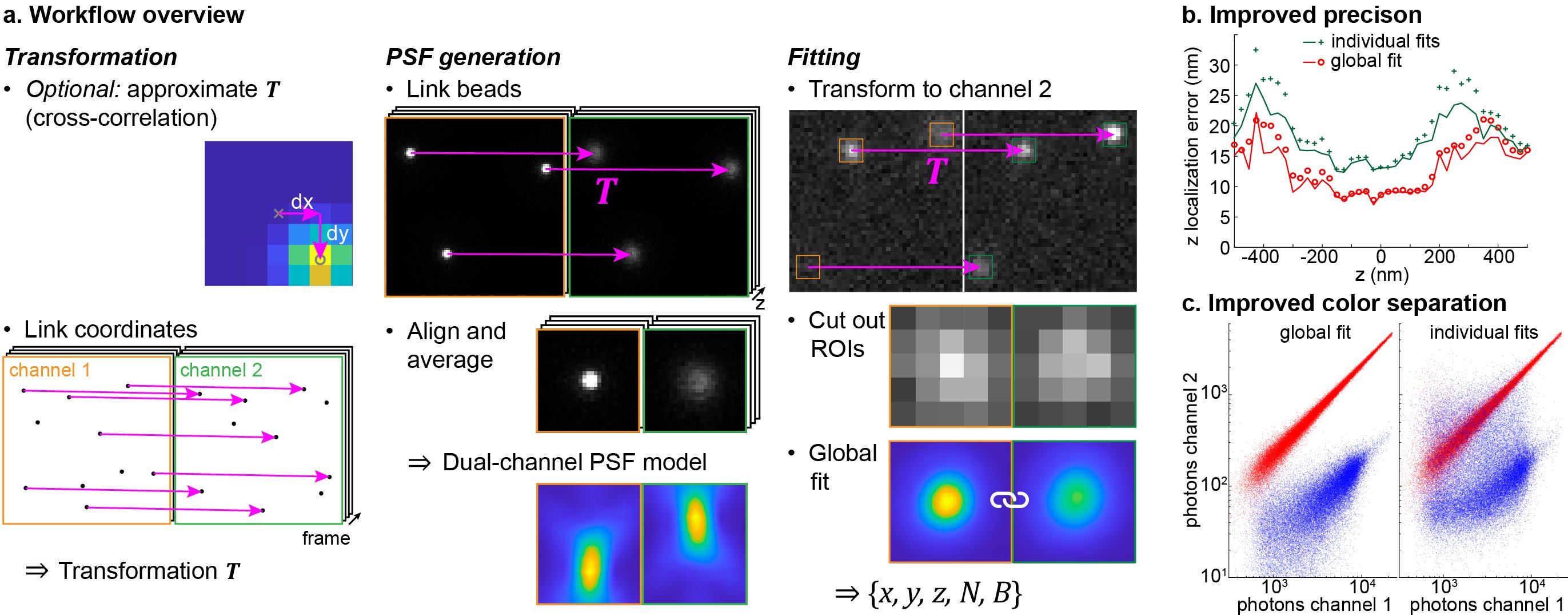
GlobLoc is a graphics processing unit (GPU) based global fitting algorithm with flexible PSF modeling and parameter sharing, to extract maximum information from multi-channel single molecule data. Global fitting can substantially improve the 3D localization precision for biplane and 4Pi SMLM and color assignment for ratiometric multicolor imaging. The fitting speeds achieve ~35,000 fits/s on a standard GPU (NVIDIA RTX3090) for regions of interest (ROI) with a size of 13×13 pixels.
GlobLoc_matlab
-
Matlab R2019a or newer
- Curve Fitting Toolbox
- Optimization Toolbox
-
The GPU fitter requires:
- Microsoft Windows 7 or newer, 64-bit
- CUDA capable graphics card, minimum Compute Capability 6.1
- CUDA 10.1 compatible graphics driver (for GeForce products 471.41 or later)
-
The CPU version runs on macOS and Microsoft Windows 7 or newer, 64-bit
GlobLoc_python
-
python 3.8 or newer
pip install numba
more details on https://numba.readthedocs.io/en/stable/user/installing.html
pip install tqdm
more details on https://github.com/tqdm/tqdm#installation
-
The GPU fitter requires:
- Microsoft Windows 7 or newer, 64-bit
- CUDA capable graphics card, minimum Compute Capability 6.1
- CUDA 10.1 compatible graphics driver (for GeForce products 471.41 or later)
-
The CPU version runs on macOS and Microsoft Windows 7 or newer, 64-bit
Examples code are avalible in file Example_GlobalFit_4Pi.m, Example_GlobalFit_biplane.m, Example_GlobalFit_Gauss.m, Example_GlobalFit_Ratiometric.m. The required test data for the demo code can be found in the folder by following this link. GlobLoc has been fully integrated in fit3Dcspline plugin of SMAP.
Examples code are avalible in file example_4pi.py, example_biplane.py, example_gauss.py, example_ratiometric.py.The required test data for the demo code can be found in the folder by following this link. The results of the example can be found in the folder output.
A full instruction guide can be found in the supplementary material (Tutorial of globFit.pdf) of the paper.
For any questions / comments about this software, please contact Li Lab.
Copyright (c) 2021 Li Lab, Southern University of Science and Technology, Shenzhen &Ries Lab, European Molecular Biology Laboratory, Heidelberg.
If you use Global to process your data, please, cite our paper:
- Yiming Li, Wei Shi, Sheng Liu, Ivana Cavka, Yu-Le Wu, Ulf Matti, Decheng Wu, Simone Koehler, Jonas Ries. Global fitting for high-accuracy multi-channel single-molecule localization. Nat. Commun. 13, 3133 (2022).