-
Notifications
You must be signed in to change notification settings - Fork 19
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Specify disulfide bridges during parameter file generation #368
Comments
|
hi @cespos - thanks for the post. We will discuss your suggestion and post an update (sometime next month.) |
|
Great, thanks! |
|
Hi @cespos. Thanks for your query, this is really helpful. The As you say, ideally it would be possible to pass a fully custom config file, or modify it as you can do for a I'll have a think at how to best implement this, then run some suggestions by you. Cheers. |
|
hi @cespos, you can also use the import BioSimSpace as BSS
pdb = BSS.IO.readMolecules('4LYT_Fixed.pdb')
# define custom leap script except for ff14SB sourcing
custom_commands = ['4lyt = loadPDB leap.pdb', # note the use of 'leap.pdb'
'bond 4lyt.6.SG 4lyt.127.SG',
'bond 4lyt.30.SG 4lyt.115.SG',
'bond 4lyt.64.SG 4lyt.80.SG',
'bond 4lyt.76.SG 4lyt.94.SG',
'saveamberparm 4lyt custom.prm7 custom.rst7']
parm = BSS.Parameters.ff14SB(pdb.getMolecule(0), leap_commands=custom_commands)
# load the custom molecule directly from working dir
parm_mol = BSS.IO.readMolecules(f'{parm.workDir()}/custom.*')
BSS.IO.saveMolecules('4LYT_parm', parm_mol, ['rst7', 'prm7'])I run the following to check that the S-S bonds are present in the topology: $ cpptraj
> parm 4LYT_parm.prm7
> bondinfo :6@SG
# Mask [:6@SG] corresponds to 1 atoms.
#Bnd RK REQ Atom1 Atom2 A1 A2 T1 T2
1006 227.00 1.810 :6@CB :6@SG 96 99 2C S
1007 166.00 2.038 :6@SG :127@SG 99 1914 S Syou can see that SG in residue 6 is bonded to SG in residue 127 |
|
We should easily be able to detect the present of disulphide bonds using Sire, then generate the correct import BioSimSpace as BSS
from sire.legacy.Mol import Connectivity
from sire.legacy.Mol import Element
S = Element(S)
# Load the molecule.
mol = BSS.IO.readMolecules("4LYT_Fixed.pdb")[0]
# Generate the connectivity (bonding) object for the Sire molecule, then
# add this as a property.
connectivity = Connectivity(mol._sire_object)
mol._sire_object = mol._sire_object.edit().setProperty("connectivity", connectivity).molecule().commit()
# Get the bonds associated with the Sire molecule.
bonds = mol._sire_object.bonds()
# Create a list to store our disulphide bonds.
disulphides = []
# Loop over the bonds and store the disulphides.
for bond in bonds:
if bond.atom0().element() == S and bond.atom1().element() == S:
disulphides.append(bond)We then just need to loop over the |
|
Okay, I've got this working on the feature-disulphide branch. The actual function that generates the records is here, which is basically the same as above with the addition of generating the record string for each bond. The one thing that I'm concerned about is that determining the bonds via the |
|
I had a quick Google and found a file with the same name in an AMBER tutorial here. Using this, I only generate two bond records, i.e.: import BioSimSpace as BSS
m = BSS.IO.readMolecules("4LYT_Fixed.pdb")[0]
BSS.Parameters.Protocol._protocol._get_disulphide_bonds(m._sire_object)
['bond mol.6.SG mol.127.SG', 'bond mol.64.SG mol.80.SG']Perhaps this file is different, or maybe the |
|
I can get it to work by tuning the tolerance, but since the bond hunter is based on covalent bonds, then I also need to tweak the maximum bond limit. I'll see what settings are most reliable. In practice, it might be best to add a new bond hunter specifically for disulphide bridges. |
|
Could Sire parse |
|
Thanks, that is also an option. To date we haven't parsed those records since they are often missing or incomplete, so it's hard to detect when things are wrong. Since we rely on a well formatted PDB for paramterisation in general, I think it might be best to assume well formatted |
|
Ah, didn't realise that Sire also has a |
|
I've made some edits to Sire to make the bond searching more flexible. The user can now specify the maximum search distance and a tolerance to use when comparing equilibrium bond radii for determining bonds. I've then tuned the parameters so that it works well for a few test systems. I'll let some other collaborators test this against some of their input in order to work out where the edge cases are. It should be easy enough to fall back on the Cheers. |
|
Hi @lohedges, Sorry I saw this only now. I will test this functionality asap and let you know how it works for me. |
|
Yes, that's the correct branch. The way in which I search for disulphides is quite flexible to allow for non-ideal configurations. It would be good to know if it's too flexible, e.g. picking up things that it shouldn't. I'm also working on some improved |
|
Related to other discussions here and here, I've made some improvements to the In doing so, I have switched to writing PDBv3 compliant One issue that I have discovered, which would also be a problem for the previous approach, is that the atom numbers used to generate the Cheers. |
|
Oh, and I have now added a |
|
Okay, I've realised that I can match the atom coordinates from the |
|
Hi @lohedges, |
|
Assuming that you have a correctly formatted PDB file, the code should now autodetect disulphide bonds and add them to the
See here for the documentation for |
|
There's also a unit test here that checks that the parameterisation works and that the resulting AMBER topology does contain the required disulphide bonds. |
|
The current |
|
Weird, I installed it today following the instructions. I'll figure it out and get back to you. |
|
It's possible that it's pulling in an older version because it doesn't want to change the version of Sire in your environment. To force it to use the latest you can do: Or create an environment directly (which is recommended): |
|
This worked: |
|
The commands are now working... at least partially. tleap however failed. The problem now is that tleap residues/atoms numbering should start from 1 and should be continuous. When I checked the leap.pdb in the temporary work_dir, I found that residues numbers were not continuous (no residue 16): As a consequence, the bond specifications are not working. A similar issue will occur when proteins have multiple chains and there are disulfide bridges involving the different chains. The solution for those cases is to first run tleap to generate a processed pdb file (with continuous residues/atoms numbers). Then, check whether there are any disulfide bridges and generate the tleap bond commands. Finally, run tleap again to parametrize the protein with disulfide bonds. See also this. |
|
This is because your original input PDB file does not have continuous residue numbers. With BioSimSpace we make the assumption that the user input file is the source of truth and has been formatted appropriately. As such, we write back things like residue numbers as is, since the user might want to cross-reference things later down a pipeline. (We don't change things behind the users back.) If this is a common issue with PDB input files, then could you provide some examples (particularly the multiple chain issue you mention). It might be possible to fix the files, i.e. make things continuous, then remap to the original numbering after the parameterisation is complete. (We already make use of a compatibility function that maps the output topology of |
|
Hi @lohedges, Sorry again for taking a long time to follow up. I was thinking... |
|
Thanks for this, I'll look into it next week. |
Sorry, perhaps I'm misunderstanding something here, the bonds do you mean use the residue indices instead of the number used in the original PDB, rather than the name? If |
|
I mean, the current bond record uses the residue number and name from the PDB, so just the name part should be replaced with the index? |
|
Sorry if I was unclear. But yes it should be an easy fix :) So, in the LEAP file in the disulfide bond definitions, instead of specifying residue numbers read from the PDB (e.g. To be honest, I did not test this out on the complicated cases that I sent you, but I can try to do that later before you make any changes! Thanks! |
|
Great, that would be really helpful. |
|
Hi Lester! If you use the function Notice that the list has "duplicate" bond specifications. However, these are not really duplicates but are due to the presence of two chains with equal residue numbering. I tried to parametrize this protein with Here is the tleap configuration file that I used: Notice also that using the indices, there are no duplicate bond definitions. So... it should be an easy fix to the I tested also the 5Y42 case and worked as well. I am attaching here prepared PDB files and tleap configuration files (with txt extension). Best, |
|
Thanks for this, I'll create a new feature branch and see if I can get it working. For the purposes of writing the file for |
|
This is now implemented here. Note that using your files directly, I can only get the |
|
Hi! It looks like it worked!! All my examples run through! I think we can close this issue and merge to main branch. |
|
Fantastic. Thanks for confirming. I have another external collaborator who is also testing the update against their benchmark set. I'll close this when they are happy that things are still working. Thanks again for your input. It turns out that the solution was much simpler than I originally thought. |
|
Thanks to you! |





Is your feature request related to a problem? Please describe.
I have tried to prepare a system containing 4 disulfide bridges, however SS-bridge parameters were not in the output parameter file. When generating the parameter file using tleap, it is necessary to specify the SS-bridges explicitly in the following format after the pdb has been loaded.
With BSS, it is not possible to insert these lines in the leap configuration file because although there is a leap_commands option, extra lines are written before the system is loaded.
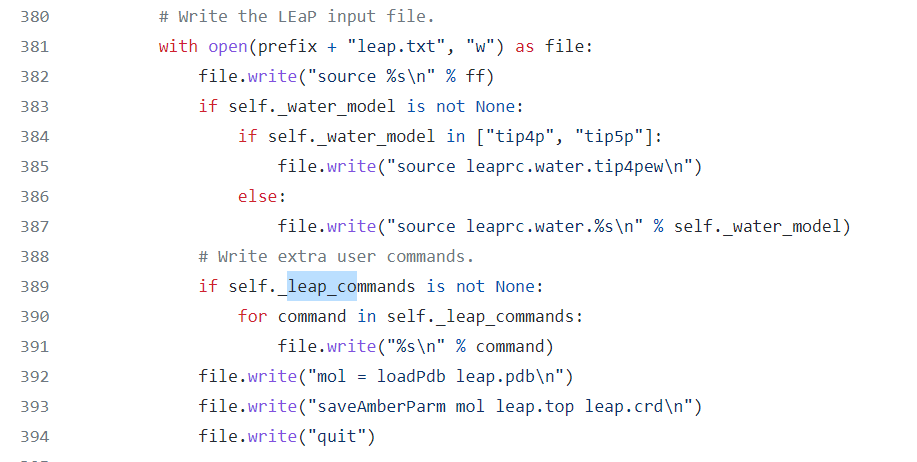
Describe the solution you'd like
Would it be possible to have a leap_insert_flag, to decide where to insert the additional "leap_commands"?
Something like "head", "pre-load", "post-load", "tail"?
Describe alternatives you've considered
Or alternatively, would it be possible to have the option to specify your own tleap configuration file (as it is possible for MD configuration files)? ...Maybe this alternative would be even more useful because will allow for more flexibility.
Additional context
Add any other context or screenshots about the feature request here.
The text was updated successfully, but these errors were encountered: