-
Notifications
You must be signed in to change notification settings - Fork 22
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Feature request: paired forest/risk-of-bias plot #102
Comments
|
Yes, happy to help and/or for my code to be used. A couple early thoughts:
|
|
Looking at the figure a little more, here's how I think we could build it:
I don't think it's possible to align the plots correctly using patchwork if they're three separate objects, but I could be wrong |
|
IMHO the "statistical tests as rows" design is kind of distracting. It should work no problem, but I wonder if there's a better place to put them |
|
Thanks for your feedback/thoughts @rdboyes! I completely agree with points 1/2 - this all seems very doable. I think one of the most time-consuming bits will actually be parsing the output of functions like Re: point 3, what is the advantage of producing three different plots rather than just having one, with a logical "show" variable which sets the point colour to "transparent" if a point does not need to be plotted for that row? This would also serve to negate the current Using this approach, I've managed to make a start by adapting your inital code, just as an initial proof of concept (the two-dotted ROB image on the right is all one My adapted version of your code is available here: https://gist.github.com/mcguinlu/621461bb511a8c73534c07df139e1220. Line 57 defines the "show" variable, while Lines 101-108 create the ROB plot. Finally, while I think we'll have to produce an option to produce plots with "statistical tests as rows" design, as this is what users most frequently wish to see, we could also offer an alternative opinionated version of the plot - for this version, out of curiousity, where would you put the results of the statistical tests instead? |
|
Sorry, I didn't see this response until just now. I've implemented a pretty good version of the required function that processes output from metafor::rma() automatically. It needs expanding to increase the number of formats that it is compatible with, but I think its a good baseline. I've submitted it as a pull request. |
|
And re: "why not just have one ggplot object?" - we agree on this, I just phrased it in a confusing way. |
|
Thanks for the tag @mcguinlu, looks like you guys have this covered - the plots look really great! |
|
The inital version of this function is now in the Functionality
Documentation
Test
Look and feel
Misc To Do
|
|
@kylehamilton - just saw you fork |
|
Amazing watching this unfold! Top of my feature list is flexible grouping and ordering and plotting. My current use case is to convert three custom forest plots for different outcomes into a single plot. It's such a clear way to visualise and compare effect sizes, mentally moderated by the traffic lights. |
|
@wviechtb, tagging you here as my supervisor (Julian Higgins) suggested it would be worthwhile flagging this to you, both out of interest and in case you had any thoughts on the proposed plan/our handling of the |
Don't hate on R default mono! Actually though the reason for choosing this font for the first pass is that a monospace font makes it much easier to troubleshoot horizontal alignment and this one is the only monospace font available in R by default. I'm working on accurately measuring width (and height, annoyingly) using |
Sincere apologies - I was just spoiled by the font you used in the inital NEJM figure 😄
Never realised this would be an issue - huh. And that all sounds great - thank you! I'm hoping to have some time this weekend myself to have a look at adapting your code to address some of the other functionality. Out of interest, is reading from |
I was thinking about this as well, but I'm not sure of the best way to proceed. I think it does make sense to add comprehension for MA objects to |
Just to flag that And I think having As an alternative, I'm very happy to try and develop the I guess it really depends where support for |
|
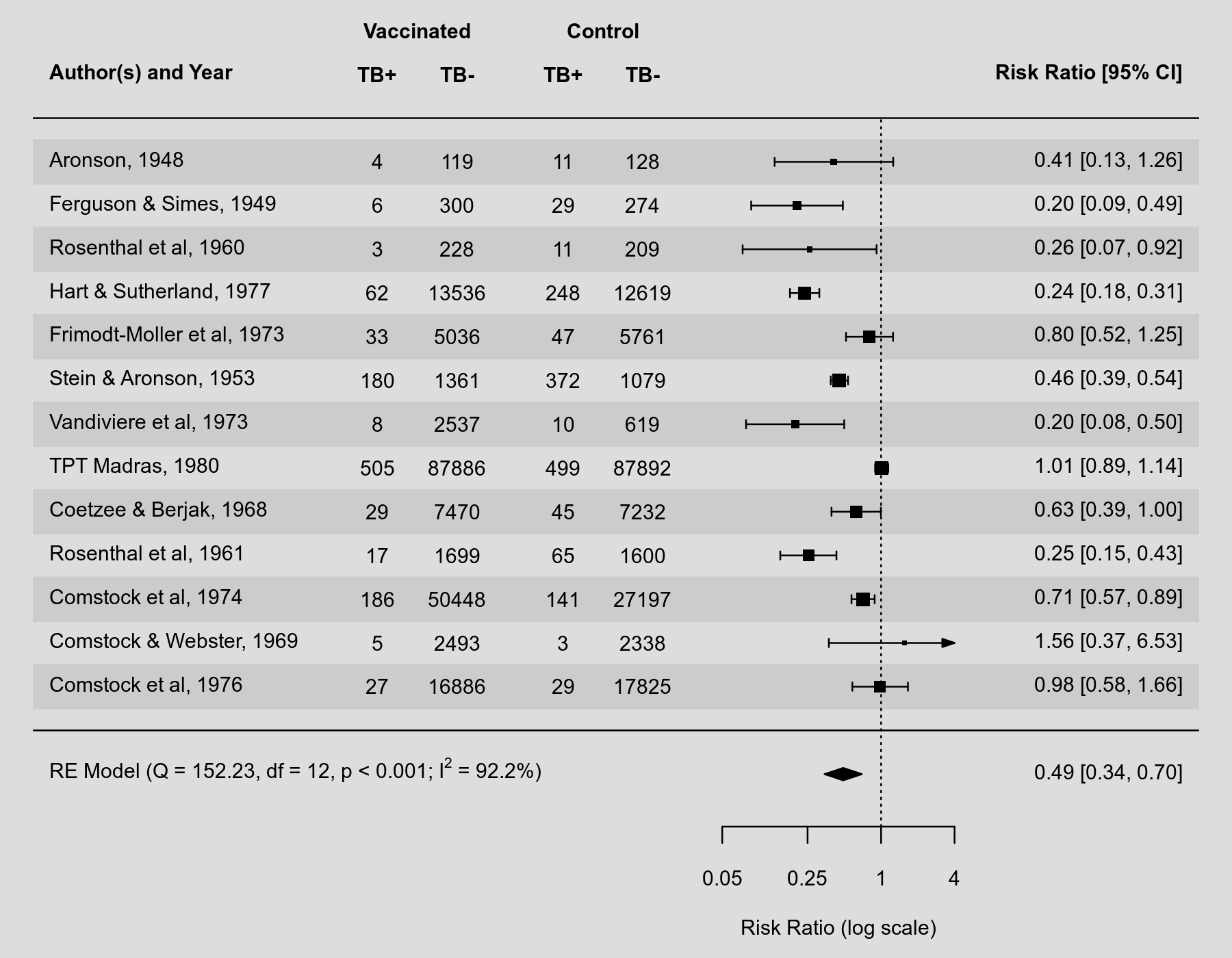
Adding some additional colored points could be done as follows. If you want them to the right of the annotations (estimates, CI), then you can use the (currently undocumented) library(metafor)
### copy BCG vaccine meta-analysis data into 'dat'
dat <- dat.bcg
dat$bias1 <- sample(1:3, nrow(dat), replace=TRUE)
dat$bias2 <- sample(1:3, nrow(dat), replace=TRUE)
dat$bias3 <- sample(1:3, nrow(dat), replace=TRUE)
dat$bias4 <- sample(1:3, nrow(dat), replace=TRUE)
### calculate log risk ratios and corresponding sampling variances (and use
### the 'slab' argument to store study labels as part of the data frame)
dat <- escalc(measure="RR", ai=tpos, bi=tneg, ci=cpos, di=cneg, data=dat,
slab=paste(author, year, sep=", "))
### fit random-effects model
res <- rma(yi, vi, data=dat)
### forest plot with extra annotations
forest(res, atransf=exp, at=log(c(.05, .25, 1, 4)), xlim=c(-16,11),
ilab=cbind(dat.bcg$tpos, dat.bcg$tneg, dat.bcg$cpos, dat.bcg$cneg),
ilab.xpos=c(-9.5,-8,-6,-4.5), cex=.75, header="Author(s) and Year",
mlab="", textpos=c(-16, 6))
op <- par(cex=.75, font=2)
text(c(-9.5,-8,-6,-4.5), 15, c("TB+", "TB-", "TB+", "TB-"))
text(c(-8.75,-5.25), 16, c("Vaccinated", "Control"))
text(7:10, 15, c("B1", "B2", "B3", "B4"))
par(op)
### add risk of bias points
cols <- c("green", "yellow", "red")
syms <- c("+", "?", "-")
points( rep(7,13), 13:1, pch=19, col=cols[dat$bias1], cex=2)
text(7, 13:1, syms[dat$bias1], cex=0.8)
points( rep(8,13), 13:1, pch=19, col=cols[dat$bias2], cex=2)
text(8, 13:1, syms[dat$bias2], cex=0.8)
points( rep(9,13), 13:1, pch=19, col=cols[dat$bias3], cex=2)
text(9, 13:1, syms[dat$bias3], cex=0.8)
points(rep(10,13), 13:1, pch=19, col=cols[dat$bias4], cex=2)
text(10, 13:1, syms[dat$bias4], cex=0.8)
### add text with Q-value, dfs, p-value, and I^2 statistic
text(-16, -1, pos=4, cex=0.75, bquote(paste("RE Model (Q = ",
.(formatC(res$QE, digits=2, format="f")), ", df = ", .(res$k - res$p),
", p = ", .(formatC(res$QEp, digits=2, format="f")), "; ", I^2, " = ",
.(formatC(res$I2, digits=1, format="f")), "%)")))This would then look like this: |
|
Thanks everyone for pitching in on this! A few updates for those watching out of interest. Having thought it over for a while now, I have decided to take the approach of being over inclusive, so that >1 option available to users, depending on what set-up they are most comfortable with. The result is that the "blobbogram" branch of
There is still much work to be done on both functions, including adding support for more risk-of-bias tools and improving the look/feel of the output of the Finally, any and all feedback/thoughts/comments are very welcome!Other notes@wviechtb - as @rdboyes - I spent a lot of time trying to work out how best to do the testing for the |
|
Very nice! I didn't really do anything with respect to |
|
Sorry to leave this so long - other projects took priority, etc. I've updated the The main thing I'm trying to avoid with this setup is duplicating my |
|
No problem at all - I've also been a bit stretched for time recently.
That all sounds great - being able to do just a little preprocessing and then pass it all off to Just to note, I have merged in your initial PR now (#109) but with the caveat that the functionality offered by |
|
Making progress on this! I've uploaded my current code in a vignette (which is absolutely not useful to anyone else yet) here: https://github.com/rdboyes/forester/blob/master/vignettes/Adding_Custom_GGPlots.Rmd but the punchline looks like: |
|
Just what I've been looking for :) |
|
Ok! A reasonably effective version of the function that does generally what we originally wanted (subsets by any column, supports all major tools and colour schemes, and depends on forester as a backend) is in my fork of robvis. I haven't made a pull request yet because I'm currently getting a lot of failed tests - not sure exactly what's going on. Here's how it looks so far though! Robins, serif, cochrane Quadas, sans, cochrane ROB2, serif, colourblind |
|
This is great @rdboyes. I'm trying to apply the vignette to my own data. At the moment you can't do ROB2 and ROBINS in the same plot. Is there a methodological reason why you should include RCTs and non-RCTs in the same meta-analysis? |
|
I don't have any intention of adding support for two different tools in the same plot, but you can probably modify the code to make that happen if it's important to you. I'd suggest just making two separate plots as a simple workaround. Your last question is outside my specific expertise, but I would say yes, there are situations where RCTs and non-RCTs target the same estimand and could be shown in the same plot, though there are many assumptions involved to make it make sense |
|
[Hi both, I'm going to respond to you two seperate comments] Hi @earcanal, Gald you are finding this amazing work by @rdboyes helpful. Re: putting RCTs and non-RCTs on the same plot, I would echo Randall's point about there being some situations where this is appropriate. However, these situations are quite rare, and adding standardised support for them in |
|
Hi both, Thanks for your thoughts on this. I'm going to keep my RCTs and case-controls as separate plots. I am going to attempt to adapt the vignette to work with |
|
@rdboyes sorry for the delay - this is all looking superb! There are a couple of small additional aesthetic tweeks I'd like to make, but I can do this myself and I think it's worth getting the function into use as is. Two things I wanted to query that I think are currently handled from with
I had a look at the test suite on your fork and it looks like everything is passing now, barring the test coverage check (which is a minor issue) and the Ubuntu devel where it can't find Very happy to review a PR whenever you have time to open one, and as always, thanks for your work on this! 😁 |
|
The whitespace is easy (he said, foolishly) - it's currently there because the axis on the left side is being drawn but is transparent. I should be able to instead not draw it with Aligning the titles is more problematic. The way the alignment currently works inside For what it's worth, personally I prefer the labels where they are over having them aligned with the table :) |
Ha, I sense a kindred spirit! But great that it is relatively easily solvable!
Ah I thought there was something else going on. Personally I think both options (where they are currently vs aligned with the table) look a bit odd - incredibly unhelpful I know, but I can't think of where I'd actually like to put them. So in absence of a personal preference, I'm going to agree with you and say leave them where they are (which happily is computationally easier too!) |
|
Great! I've fixed the whitespace issue and submitted what I have so far in a pull request. My feeling is that this function is still "development" status - not ready for prime time, but progressing towards it! |
|
Hi @rdboyes I am loving this new functionality. Have been working hard on it recently to try and get it ready for formal release. As part of this, I have some thoughts/questions:
[Of interest, rather than implementing the above yourself, it might be easier to allow users to replace the default central ggplot2 graph in a similar fashion to
Hi @paulsharpeY, Looks interesting! My plan is to get it up, running and fully supported for metafor before (maybe) branching out in other MA packages. But great to know if works 👍 |
|
FWIW, I use |
|
@mcguinlu Just noting that I've seen this and will respond when I have time (i.e. not right now) |
Yep, I can add that functionality no problem. This should be easy as well A little trickier but doable Yep, I can do that This is the trickiest one. I think the easiest way to fix this is to blank the actual title row (just colour it white) and create a fake header row in the first row Probably a good idea to quiet it down, yep I've never submitted to CRAN before, but assuming it's reasonable to get it done, I'll likely do it |
|
Progress made on a few of these issues! The newest version supports |
|
Hey @mcguinlu - I think I have a working fix for the final issue (this one)
Do you have somewhere where the data/code for that example lives already? Just so I don't have to recreate it |
|
Hi @rdboyes, this all looks amazing - hoping to have some time over the Easter weekend to run through it all properly. Re: the column heading example, essentially, I have added an additional Lines 268 to 314 in 65737f1
So having loaded that version of the function, running the following code will produce the figure I included in the comment above. Hopefully this is what you were looking for, but let me know if I can provided anything else useful! |
|
Hi @mcguinlu, this function looks very nice. Yet, I wasn't able to download it. Could you please help me? Thanks! |
|
I need making a rob_append_to_forest, but I can´t do this. |













Problem
The most recent paper on ROB2, and indeed the Cochrane Handbook itself, suggest presenting risk-of-bias information as part of a paired forest plot. Risk-of-bias assessments are intended to be result-specific, and so presenting information about the potential bias in a result alongside the main analysis helps to ensure that the results are interpreted appropriately. The paired figure from the ROB2 paper is below, for reference:
This plot is something that has been requested by other users and has been on the cards for a while:
Until recently, I was not sure how to do it, as the
metafor::forest()function is quite restrictive in terms of matching with anything else, and aligning text tables inggplot2is a pain. However, @rdboyes recently shared code for producing a beautiful forest plot to Twitter. The code is available here: https://github.com/rdboyes/Forest_plot_with_table/blob/master/create_figure.RDescribe the solution you'd like
Ideally, I'd like to add a function (
rob_blobbogram()) torobviswhich creates these table/forest-plots/risk-of-bias plots.The function would take two main inputs:
rmaobject frommetafor::rma())rob_*()functions (for consistency)Plus then some styling arguments, similar to the other
rob_*()functions.Describe alternatives you've considered
I have looked at packages like
patchworkandcowplotto see if two disctinct plots could be aligned easily. It is possible - howeverggplot2is not great at handling text/tables. So while the forest plot and risk-of-bias plots could be aligned, adding the extra tables describing the study's results proved difficult.Predicted issues
One important issue is that when previewing the plot created by @rdboyes, it looks scewy in the RStudio viewer but perfect once saved to a file. From a user interface point of view, this will cause a lot of issues as people email to say the graph looks wrong. I think it should be possible to address this by setting the dimensions of the RStudio viewer panel, but I haven't looked into it a whole lot. Perhaps @rdboyes, you might know?
Secondly, the current code for the forest-plot draws from a table rather than a meta-analysis object, and also does not include other aspects produced by the meta-analysis such as overall summaries or heterogeneity. However, by adding rows containing these, it should be possible to adapt the existing code to work.
Practical issues
@AJFOWLER - are you interested in having a go at this, as it might be a bit more interesting/challenging for you than refactoring the current code base?
@rdboyes - are you happy for us to reuse your forest plot code here/be involved in developing this functionality?
The text was updated successfully, but these errors were encountered: