-
Notifications
You must be signed in to change notification settings - Fork 0
/
Copy pathFishery Collapse extra code.Rmd
501 lines (405 loc) · 14.2 KB
/
Fishery Collapse extra code.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
---
title: "Relational Data Practice"
output: html_document
---
```{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE)
```
## R Markdown
This is an R Markdown document. Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents. For more details on using R Markdown see <http://rmarkdown.rstudio.com>.
When you click the **Knit** button a document will be generated that includes both content as well as the output of any embedded R code chunks within the document. You can embed an R code chunk like this:
```{r cars}
summary(cars)
```
## Including Plots
You can also embed plots, for example:
```{r pressure, echo=FALSE}
plot(pressure)
```
Note that the `echo = FALSE` parameter was added to the code chunk to prevent printing of the R code that generated the plot.
```{r}
library(tidyverse)
library(nycflights13)
airlines
airports
planes
weather
```
```{r}
flights2 <- flights %>%
select(year:day, hour, origin, dest, tailnum, carrier)
flights2
```
```{r}
flights2 %>%
select(-origin, -dest) %>%
left_join(airlines, by = "carrier")
```
##flights$tailnum is called a foreign key
flights$tailnum
```
```{r}
planes$tailnum
```
```{r}
fish %>%
filter(region == "Canada East Coast",
scientificname == "Gadus morhua") %>% count(tsid, areaid)
```
#from Athena's code:
```{r}
as_tibble(fish %>% distinct())
```
Plot Total Cod
```{r}
as_tibble(fish %>% filter(scientificname == "Gadus morhua") %>% select(-commonname) %>% distinct())
```
Shorten the table without cod:
```{r}
not_cod <- fish %>% filter(scientificname != "Gadus morhua")
as_tibble(not_cod)
```
```{r}
fish %>%
filter(tsid == "TCbest-MT", region == "Canada East Coast") %>%
group_by(tsyear, scientificname) %>%
summarise(total_catch = sum(tsvalue)) %>%
filter(scientificname == "Gadus morhua") %>%
ggplot(aes(tsyear, total_catch)) + geom_line() +
labs(x = "Year", y = "Total Catch or Landings (metric tons)", title = "Gadis morhua Data")
```
```{r}
fish %>%
filter(tsid == "TCbest-MT", region == "Canada East Coast") %>%
group_by(tsyear, scientificname, areaid) %>%
summarise(total_catch = sum(tsvalue, na.rm=TRUE)) %>%
filter(scientificname == "Gadus morhua") %>%
ggplot(aes(tsyear, total_catch, col = areaid)) +
geom_line() + facet_wrap(~areaid, scales = "free")
```
```{r}
fish %>%
filter(tsid == "TBbest-MT", region == "Mediterranean-Black Sea") %>%
group_by(tsyear, scientificname, areaid) %>%
summarise(total_catch = sum(tsvalue, na.rm=TRUE)) %>%
filter(scientificname == "Engraulis encrasicolus") %>%
ggplot(aes(tsyear, total_catch, col = areaid)) +
geom_line() + facet_wrap(~areaid, scales = "free")
```
#Athena's Species Richness Work
#Genna is trying to determine species variety at different years
```{r}
Australian_species_1950 <- fish %>%
group_by(tsyear, region, scientificname) %>%
filter(region == "Australia", tsyear == "1950")
```
```{r}
Australian_species_1950 %>%
summarize(region_richness = sum(n_distinct(scientificname))) %>%
summarize(total_richness = sum(region_richness))
```
```{r}
as_tibble(Australian_species_1950 %>%
group_by(scientificname))
```
#Species Richness (4) in the Australian Region in 1950
1. Nemadactylus macropterus
2. Sillago flindersi
3. Tiger flathead
4. Sea Mullet
```{r}
#athena, simple code, determine species richness in single region
fish %>%
group_by(tsyear, region, scientificname) %>%
filter(region == "Australia") %>%
summarize(region_richness = sum(n_distinct(scientificname))) %>%
summarize(total_richness = sum(region_richness)) %>%
#this determines the species richness of the area by adding all the times the areaid is duplicated
ggplot(aes(x=tsyear, y=total_richness)) +
geom_hline(yintercept=19, color='red') +
geom_hline(yintercept=1.9, color='red') +
ggtitle("Species richness in Australia") +
geom_col() +
labs(x="Year", y="Number of Species")
```
```{r}
#athena, fact checking that this max is correct
fish %>%
filter(region == "Australia") %>%
count(n_distinct(scientificname))
```
#Genna says, the number of species in a region throughout history is not synonymous with the global maxima on the bar graph.
```{r}
#Genna checking number of species found in Mediterranean-Black Sea region throughout the year range.
fish %>%
filter(region == "Mediterranean-Black Sea") %>%
count(n_distinct(scientificname))
```
```{r}
#Genna's attempt, from athena's simple code, determine species richness in single region
fish %>%
group_by(tsyear, region, scientificname) %>%
filter(region == "Mediterranean-Black Sea") %>%
summarize(region_richness = sum(n_distinct(scientificname))) %>%
summarize(total_richness = sum(region_richness)) %>%
#this determines the species richness of the area by adding all the times the areaid is duplicated
ggplot(aes(x=tsyear, y=total_richness)) +
geom_hline(yintercept=29, color='red') +
geom_hline(yintercept=2.9, color='red') +
ggtitle("Species richness in the Mediterranean-Black Sea") +
geom_col() +
labs(x="Year", y="Number of Species")
```
```{r}
as_tibble(Mediterranean_Black_Sea <- fish %>%
group_by(tsyear, region, scientificname) %>%
filter(region == "Mediterranean-Black Sea"))
Mediterranean_Black_Sea
```
```{r}
Mediterranean_Black_Sea_species_1950 <- fish %>%
group_by(tsyear, region, scientificname) %>%
filter(region == "Mediterranean-Black Sea", tsyear == "1950")
```
```{r}
Mediterranean_Black_Sea_species_1950 %>%
summarize(region_richness = sum(n_distinct(scientificname))) %>%
summarize(total_richness = sum(region_richness))
```
```{r}
as_tibble(Mediterranean_Black_Sea_species_1950 %>%
group_by(scientificname))
```
#Species Richness (3) in the Mediterranean-Black Sea Region in 1950
1. Xiphias gladius
2. Engraulis encrasicolus
3. Psetta maxima
#Blue Grenadier Fish Exploration
```{r}
fish <- ram$timeseries %>%
left_join(ram$stock, by = "stockid") %>%
left_join(ram$tsmetrics, by = c("tsid" = "tsunique"))
blue_grenadier <- fish
```
```{r}
fish %>%
filter(tsid == "SSB-MT", region == "Australia") %>%
group_by(tsyear, scientificname) %>%
summarise(total_catch = sum(tsvalue)) %>%
filter(scientificname == "Macruronus novaezelandiae") %>%
ggplot(aes(tsyear, total_catch)) + geom_line() +
labs(x = "Year", y = "Total Catch", title = "Blue Grenadier Data")
```
```{r}
fish <- ram$timeseries %>%
left_join(ram$stock, by = "stockid") %>%
left_join(ram$tsmetrics, by = c("tsid" = "tsunique"))
Chum_Salmon <- fish
```
```{r}
fish %>%
filter(tsid == "SSB-E00", region == "US Alaska (Pacific Salmon)") %>%
group_by(tsyear, scientificname) %>%
summarise(total_catch = sum(tsvalue)) %>%
filter(scientificname == "Oncorhynchus keta") %>%
ggplot(aes(tsyear, total_catch)) + geom_line() +
labs(x = "Year", y = "Total Catch", title = "Chum Salmon Data")
```
```{r}
fish <- ram$timeseries %>%
left_join(ram$stock, by = "stockid") %>%
left_join(ram$tsmetrics, by = c("tsid" = "tsunique"))
Norway_pout <- fish
```
```{r}
fish %>%
filter(tsid == "ERbest-ratio", region == "European Union") %>%
group_by(tsyear, scientificname) %>%
summarise(total_catch = sum(tsvalue)) %>%
filter(scientificname == "Trisopterus esmarkii") %>%
ggplot(aes(tsyear, total_catch)) + geom_line() +
labs(x = "Year", y = "Total Catch", title = "Norway pout")
```
# Exercise 2: Group Assignment
## Stock Collapses
We seek to replicate the temporal trend in stock declines shown in [Worm et al 2006](http://doi.org/10.1126/science.1132294):
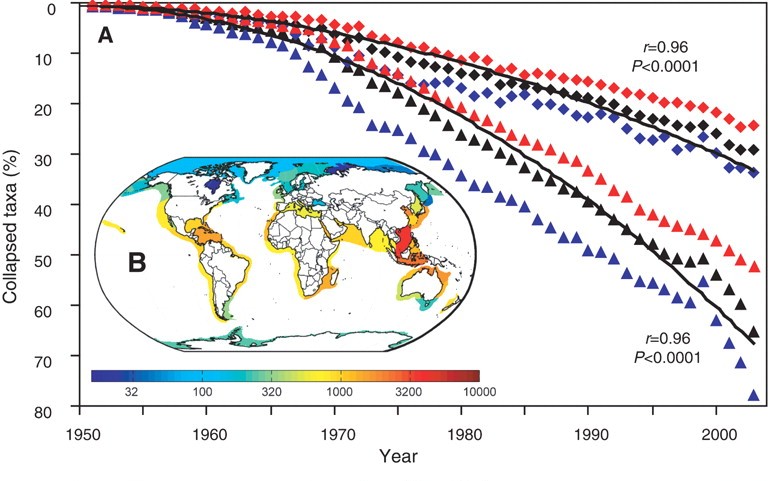
#Part 2
# from Micah: The trend line seems to be percent of total species which have collapsed by a given year, is that what we are trying to graph? Prof B says yes!
Live code from 9.30.20
```{r}
collapse <- fish %>%
filter(tsid == "TCbest-MT") %>%
group_by(tsyear, scientificname) %>%
summarise(total_catch = sum(tsvalue, na.rm = TRUE)) %>%
group_by(scientificname) %>%
mutate(current_collapse = total_catch < 0.10 * cummax(total_catch),
ever_collapsed = cumsum(current_collapse) > 0)
as_tibble(collapse)
```
```{r}
collapse <- fish %>%
filter(tsid == "TCbest-MT") %>%
group_by(stocklong.x, tsyear) %>%
summarise(total_catch = sum(tsvalue, na.rm = TRUE),
.groups = "drop_last") %>%
mutate(current_collapse = total_catch < 0.10 * cummax(total_catch),
ever_collapsed = cumsum(current_collapse) > 0)
as_tibble(collapse)
```
```{r}
collapse <- fish %>%
filter(tsid == "TCbest-MT") %>%
group_by(tsyear, stocklong.x) %>%
summarise(total_catch = sum(tsvalue, na.rm = TRUE),
.groups = "drop") %>%
mutate(current_collapse = total_catch < 0.10 * cummax(total_catch),
ever_collapsed = cumsum(current_collapse) > 0) %>%
ggplot(aes(tsyear, current_collapse)) +
geom_line()
collapse
```
```{r}
catch <- fish
as_tibble(catch %>% count(tsyear))
```
#Note from Professor B on 10.5.20
"#catch %>% count(tsyear)
will keep the tsyear column and give you n as the number of rows of data that year. and you know each row is a unique species."
Code not working
```{r}
#collapse %>%
#count(current_collapse)
```
```{r}
catch %>% group_by(scientificname) %>% summarize(n_years = n())
```
```{r}
as_tibble(catch %>% count(scientificname))
```
#Perhaps use continuous data from 1950-2003
results in denominator of 101 species?
```{r}
species1950 = unique(fish %>% filter(tsyear == 1950, tsid == "TCbest-MT") %>% select(scientificname))
length(species1950$scientificname)
```
```{r}
as_tibble(fish %>% filter(tsyear == 1950, tsid == "TCbest-MT") %>% count(scientificname))
```
```{r}
#collapse %>%
#group_by(tsyear) %>% summarise(total_collapse_species = sum(current_collapse))
```
```{r}
Total_Number_of_Species_per_year <- fish %>% count(tsyear)
as_tibble(Total_Number_of_Species_per_year)
```
```{r}
#For all taxa:
ever_collapsed <- collapse
collapse <- fish %>%
filter(tsid == "TCbest-MT") %>%
group_by(tsyear, scientificname) %>%
summarise(total_catch = sum(tsvalue, na.rm = TRUE)) %>%
group_by(scientificname) %>%
mutate(current_collapse = total_catch < 0.10 * cummax(total_catch), ever_collapsed = cumsum(current_collapse) > 0)
collapse
#Has Gadus morhua collapsed? If so, is it currently collapsed?
percentcollapse <- collapse %>%
group_by(scientificname, tsyear) %>%
summarise(total_collapse_species = sum(current_collapse))
percentever <- collapse %>%
group_by(tsyear) %>%
summarise(total_collapse_ever = sum(ever_collapsed))
taxachart <- ggplot() + geom_point(data = percentever, aes(x = tsyear, y = total_collapse_ever)) + xlab("Year") + ylab("Collapse Taxa (%)") + ylim(100, 0)
taxachart
```
```{r}
taxachart <- ggplot() + geom_point(data = percentcollapse, aes(x = tsyear, y = total_collapse_species)) + geom_point(data = percentever, aes(x = tsyear, y = total_collapse_ever)) + xlab("Year") + ylab("Collapse Taxa (%)") + ylim(100, 0)
as_tibble(percentcollapse)
```
##
scientificname
<chr>
tsyear
<dbl>
total_collapse_species
<int>
Anarhichas lupus 2009 1
Anarhichas lupus 2010 1
Anarhichas lupus 2011 1
Anarhichas lupus 2012 1
Anarhichas lupus 2013 1
Anarhichas lupus 2014 1
#Code from Prof B
```{r}
collapse <- fish %>%
filter(tsid == "TCbest-MT") %>%
group_by(stocklong.x, tsyear) %>%
summarise(total_catch = sum(tsvalue, na.rm = TRUE),
.groups = "drop_last") %>%
mutate(current_collapse = total_catch < 0.10 * cummax(total_catch),
ever_collapsed = cumsum(current_collapse) > 0)
```
#Code adapted from Slack Classmate Share
```{r}
catch <- fish %>%
filter(tsid == 'TCbest-MT') %>%
group_by(tsyear, scientificname) %>%
summarise(total_catch = sum(tsvalue, na.rm = T)) %>%
group_by(scientificname) %>%
mutate(current_tot = cummax(total_catch), current_collapse = total_catch < 0.1 * current_tot, ever_collapsed = cumsum(current_collapse) > 0)
#n <- length(unique(catch$scientificname))
years <- 1950:2003
n <- nrow(catch %>%
group_by(scientificname, tsyear) %>%
filter(all(tsyear %in% years)) %>%
group_by(scientificname) %>%
count() %>%
filter(n == length(years))) #101 species
catch%>%
filter(tsyear %in% years) %>%
group_by(tsyear) %>%
summarise(collapse = sum(current_collapse),
ever_collapsed = sum(ever_collapsed), na.rm = TRUE,
.groups = 'drop') %>%
group_by(tsyear) %>%
mutate(collapsed_taxa = 100.0 * collapse / n) %>%
ungroup() %>%
mutate(collapsed_taxa_cum = 100.0 * ever_collapsed / n) %>%
select(tsyear, collapsed_taxa, collapsed_taxa_cum) %>%
melt(id.vars = 'tsyear') %>%
ggplot(aes(x = tsyear, y = value, col = variable)) + geom_line() + scale_y_reverse() + ylab('Collapsed taxa (%)') + labs(x = "Year", y = "Collapsed taxa (%)", title = "Global Fishery Collapse")
```
#We want to see the percent collapsed_taxa_cum in 2010. (Code doesn't work 10.16.20)
```{r}
#catch %>%
#filter(tsid == 'TC-Best-MT') %>%
#summarise(cum_collapsed_taxa)
```
#Currently collapsed vs. ever collapsed fish species
```{r}
as_tibble(fish %>%
filter(tsid == 'TCbest-MT') %>%
group_by(tsyear, scientificname) %>%
summarise(total_catch = sum(tsvalue, na.rm = T)) %>%
group_by(scientificname) %>%
mutate(current_tot = cummax(total_catch), current_collapse = total_catch < 0.1 * current_tot, ever_collapsed = cumsum(current_collapse) > 0))
```
Side Note: You may notice the `lapply` function above. This function applies a given function
(in this case "read_excel") to all elements in a vector or list. This is the same
as writing out read_excel for all the sheets contained in our file, or writing
a for loop `for(i in 1:length(sheets)){read_excel(sheets[i])}`. These are very powerful
functions we will learn more about later. For now, it's enough to recognize why we
have used it here. You can find more info in [Chapter 21 of the R4ds
book](http://r4ds.had.co.nz/iteration.html).
# Exercise 1: Investigating the North-Atlantic Cod
Now we are ready to dive into our data. First, We seek to replicate the following
figure from the Millenium Ecosystem Assessment Project using the RAM data.

**How does your graph compare to the one presented above?**
------
# Exercise 2: Group Assignment
## Stock Collapses
We seek to replicate the temporal trend in stock declines shown in [Worm et al 2006](http://doi.org/10.1126/science.1132294):
